iCAZyGFADB contains seven contents: Home, CAZyme, Tools, Download, Links, Help, Register/Login.The home page also includes five parts: iCAZyGFADB’s introduction, Data Summary, Search, Submit, What's new & Cite us

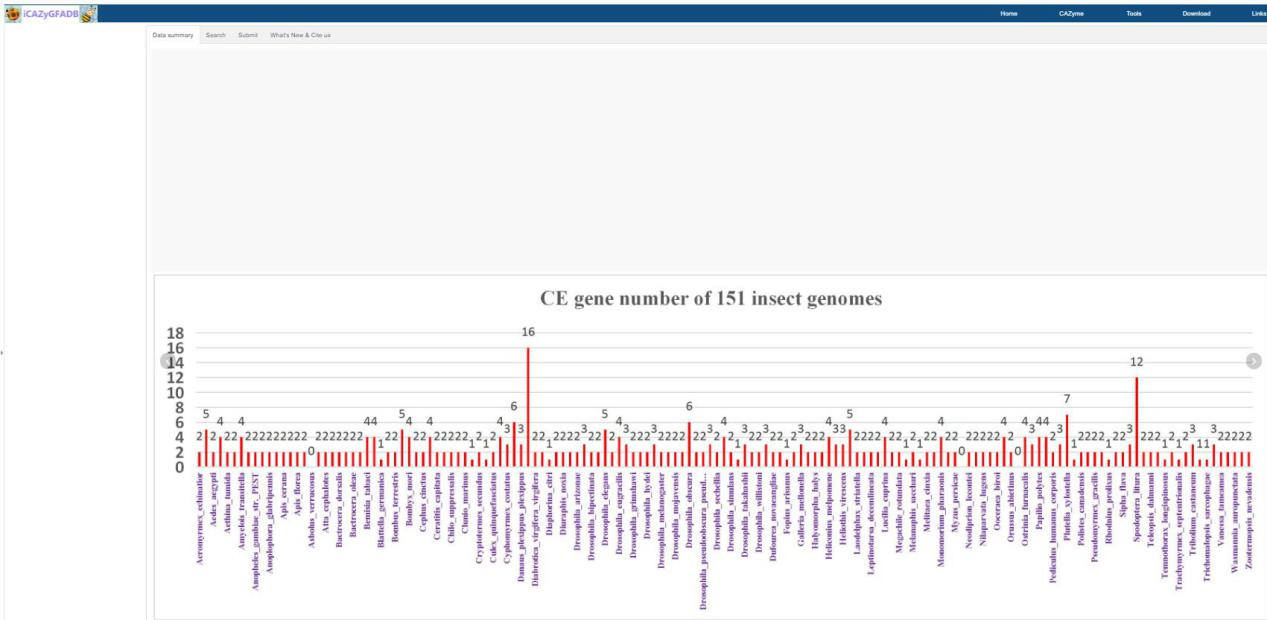
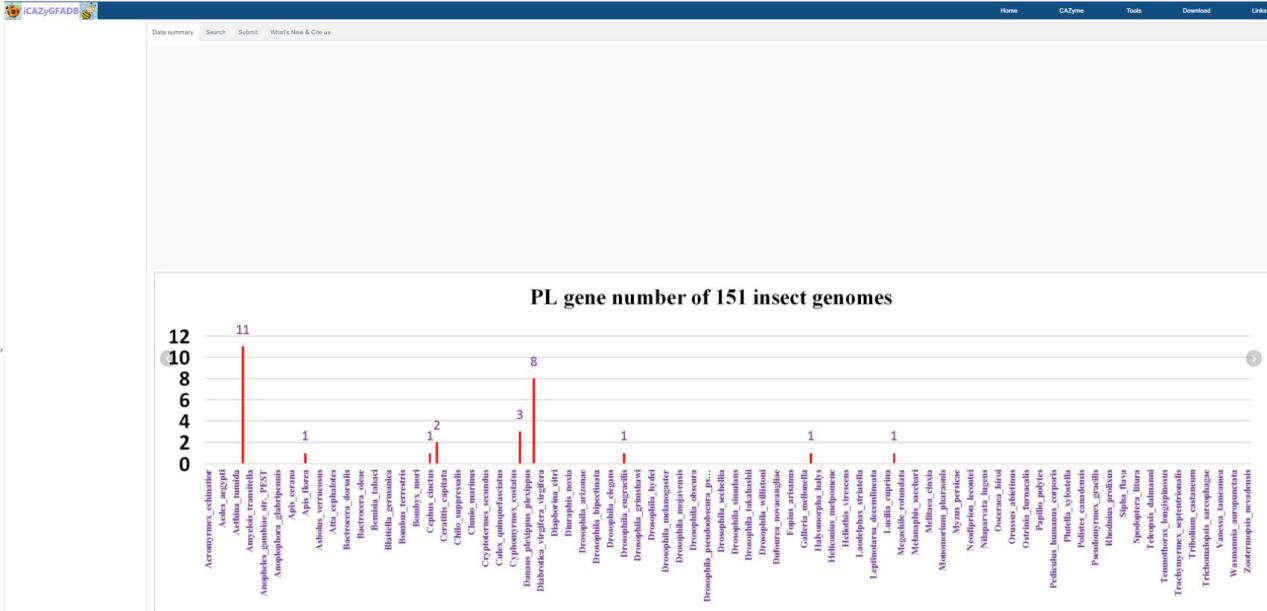
In this part, the quantitative distribution of AAs, CBMs, CEs, GHs, GTs and PLs gene family in 151 insect genomes is shown in the form of rose chart and histogram.Users can click the rose chart to zoom in and see the number of members of each AAs, CBMs, CEs, GHs, GTs and PLs gene family of each species in the high-definition map




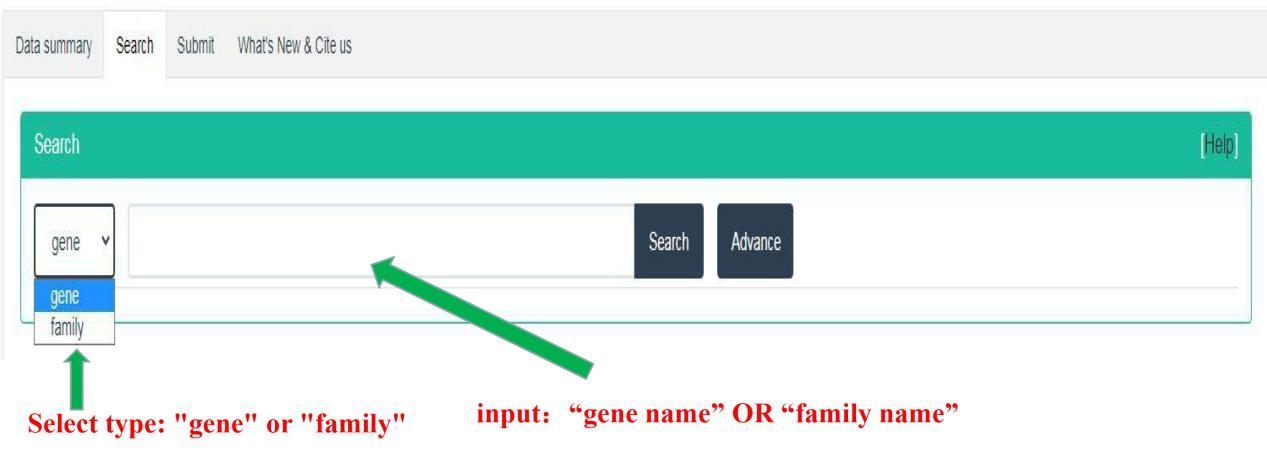
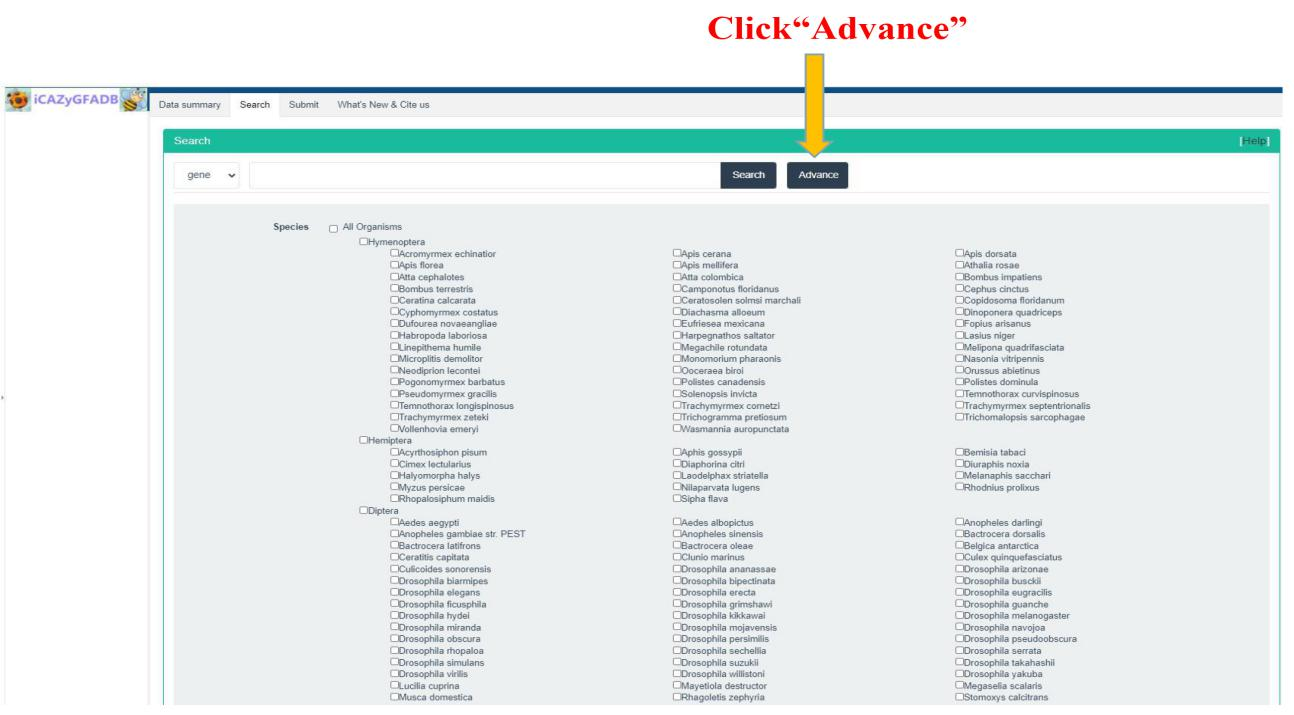
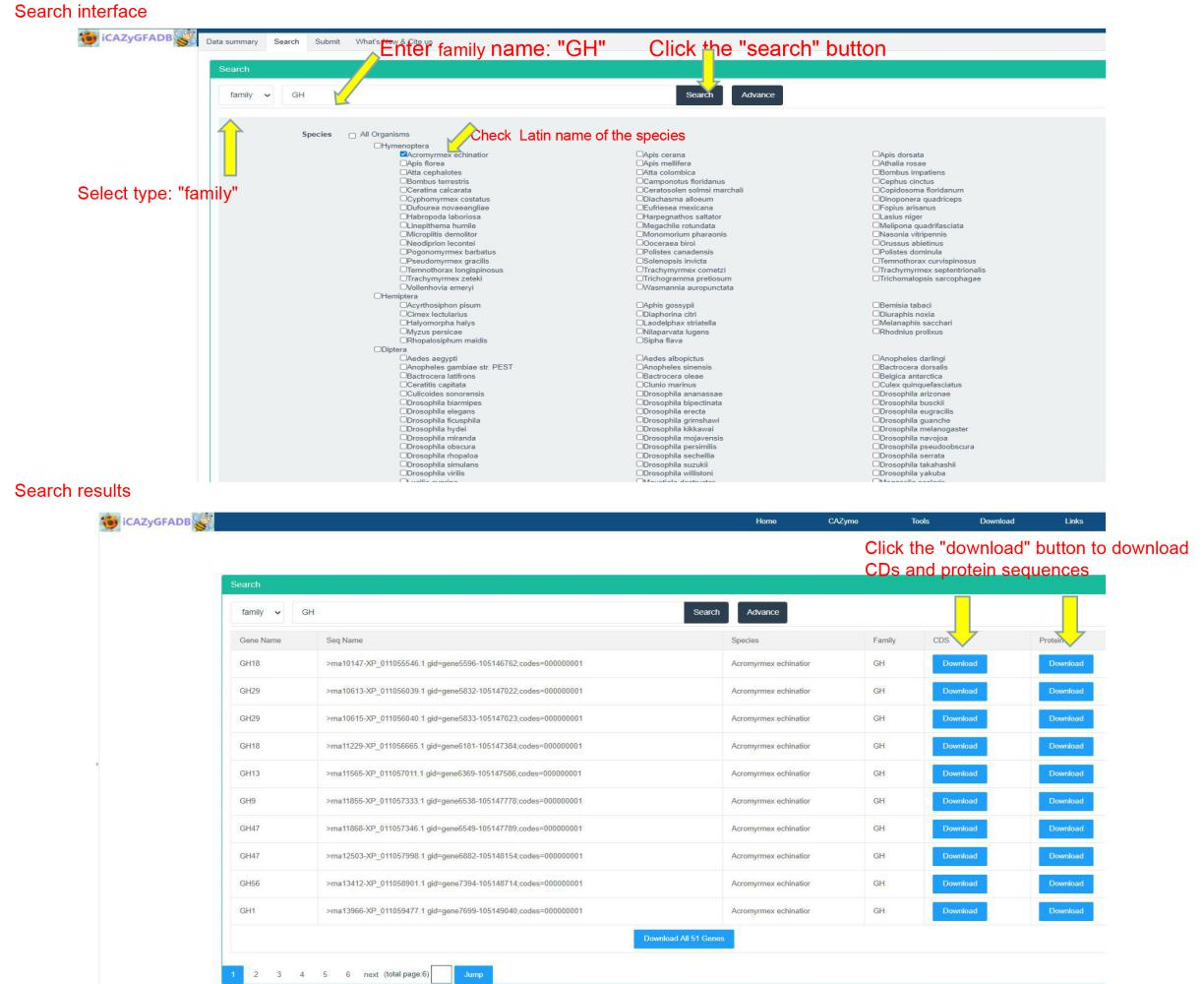
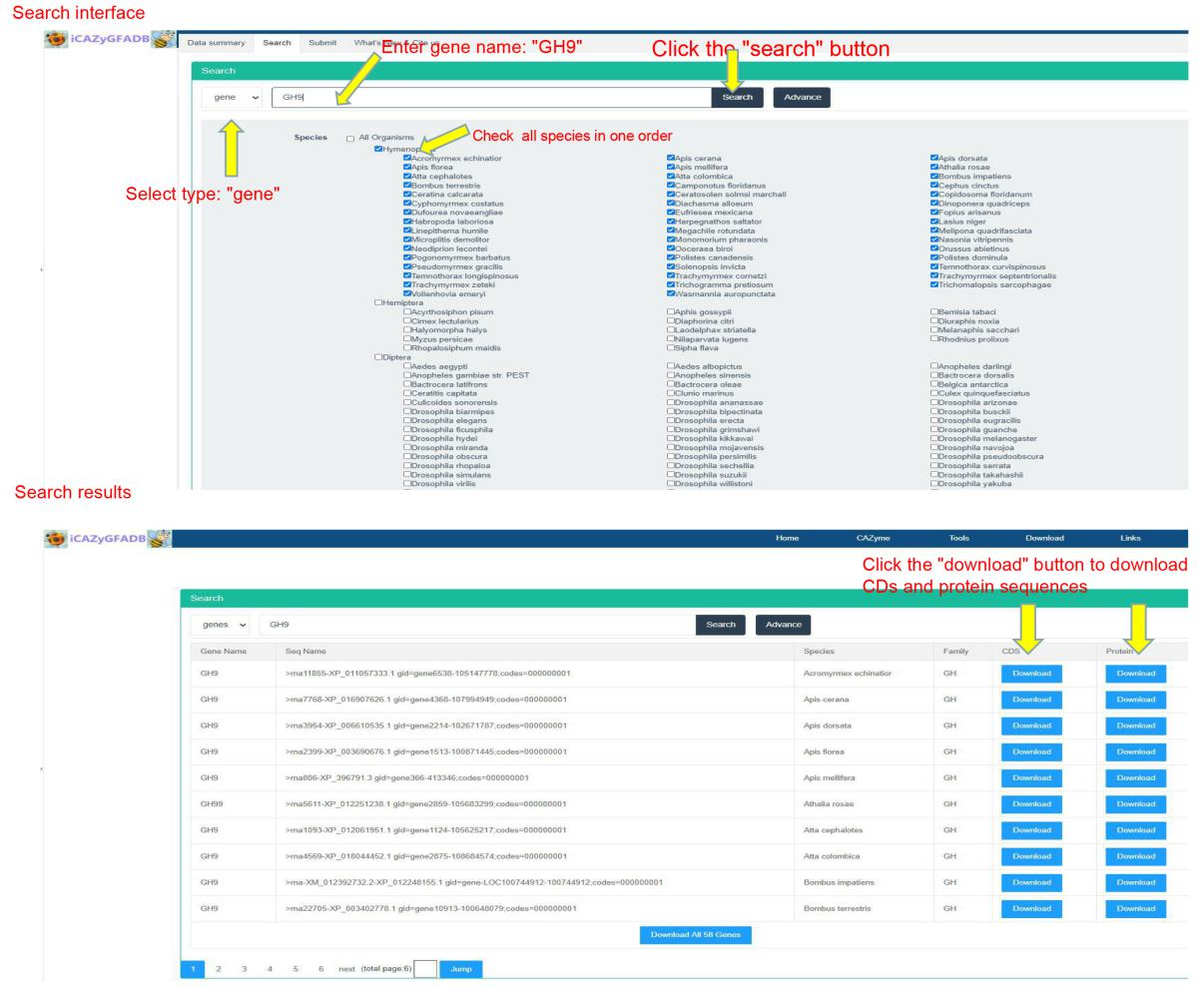
This part is to search and query the members of CAZyme gene family in 151 insect genomes from two types: gene and family. It can be further selected from different orders and species for retrieval and query by click Advance button.




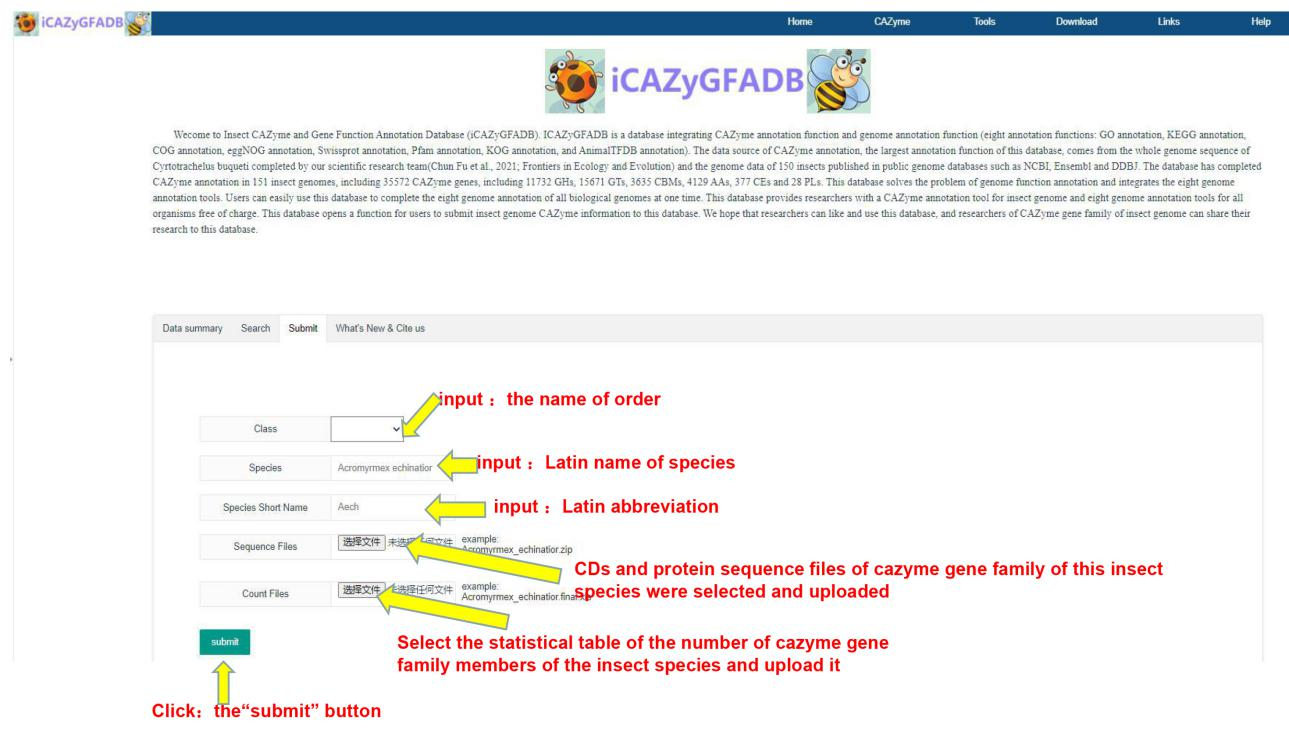
This website has developed a function that users can submit their own research data of insect CAZyme gene family to the website. Users can fill in the family, Latin name, Latin abbreviation, sequences (CDS and protein sequences) of all members of CAZyme gene family and the statistical table of the numbers of CAZyme gene family according to the data submission requirements of the website. The website administrator reviews the data submitted by users in the background, and the approved data will be updated on the download page of the database website for all users to use
This part is the design description of the database website version, the contact information of the developer and the description that users need to quote the published papers of the database when using the data and the annotation tools in the database.
CAZyme annotation tool is a user-friendly tool based on three methods: DIAMOND, HMMER and Hotpep. With reference to CAZy database, dbCAN and CAZyme peptides, CAZyme annotation tool can annotate all protein sequences in eukaryotic genome, prokaryotic genome sequences and gene sequences in Metagenome to CAZyme gene family. When using CAZyme annotation, users need to select three methods to annotate at the same time. The first step is to input the job name (less than 5 characters) and email address. The second step is to select the sequence type and annotation method. The third step is to upload the FASTA file of the annotation sequence. The fourth step is to submit.

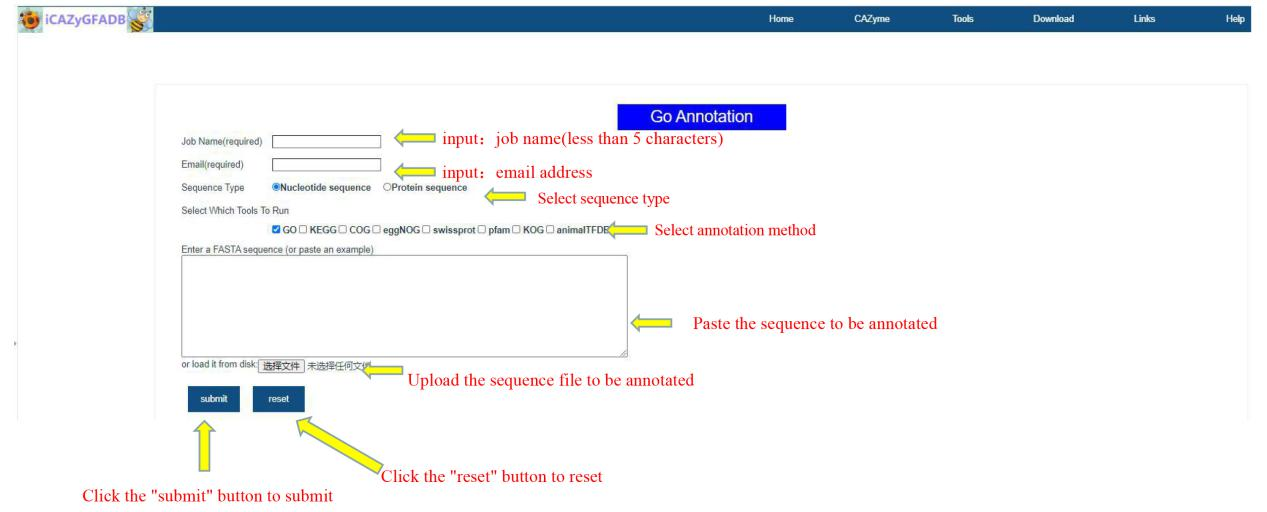
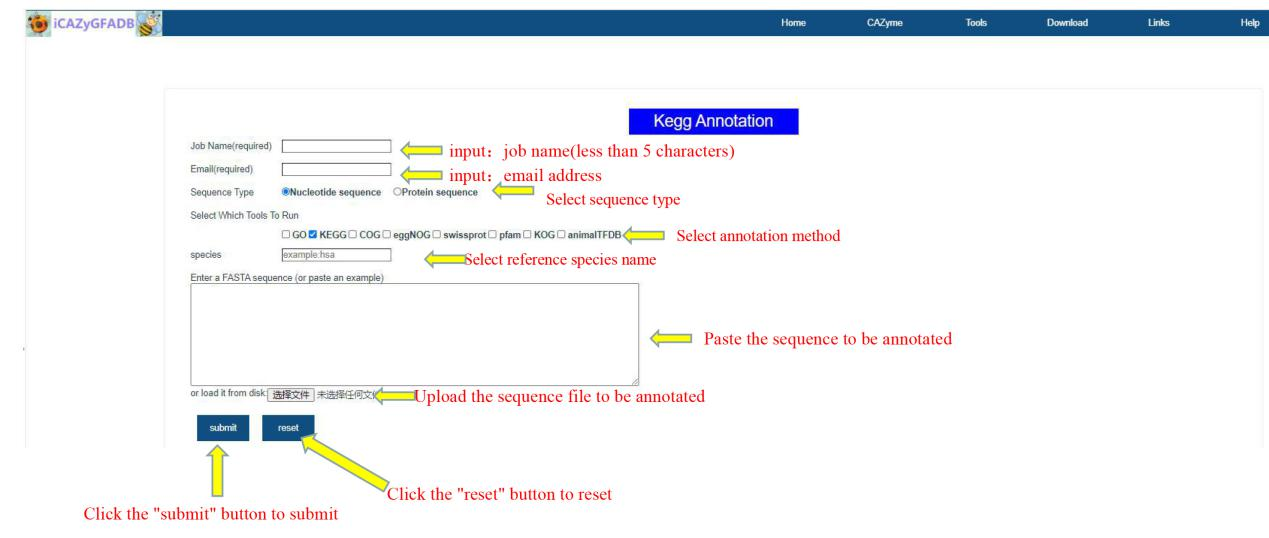
This part includes 9 gene function annotation tools, including GO annotation, KEGG annotation, COG annotation, EggNOG annotation, Swissprot annotation, Pfam annotation, KOG annotation, AnimalTFDB annotation and the above 8 simultaneous annotations.

The first step is to input the job name (less than 5 characters) and email address. The second step is to select the sequence type(Nucleotide sequence OR Protein sequence) and annotation method. The third step is to upload the FASTA file of the annotation sequence. The fourth step is to submit.
The first step is to input the job name (less than 5 characters) and email address. The second step is to select the sequence type(Nucleotide sequence OR Protein sequence), annotation method and reference species. The third step is to upload the FASTA file of the annotation sequence. The fourth step is to submit.
The first step is to input the job name (less than 5 characters) and email address. The second step is to select the sequence type(Nucleotide sequence OR Protein sequence) and annotation method. The third step is to upload the FASTA file of the annotation sequence. The fourth step is to submit.

The first step is to input the job name (less than 5 characters) and email address. The second step is to select the sequence type(Nucleotide sequence OR Protein sequence) and annotation method. The third step is to upload the FASTA file of the annotation sequence. The fourth step is to submit.
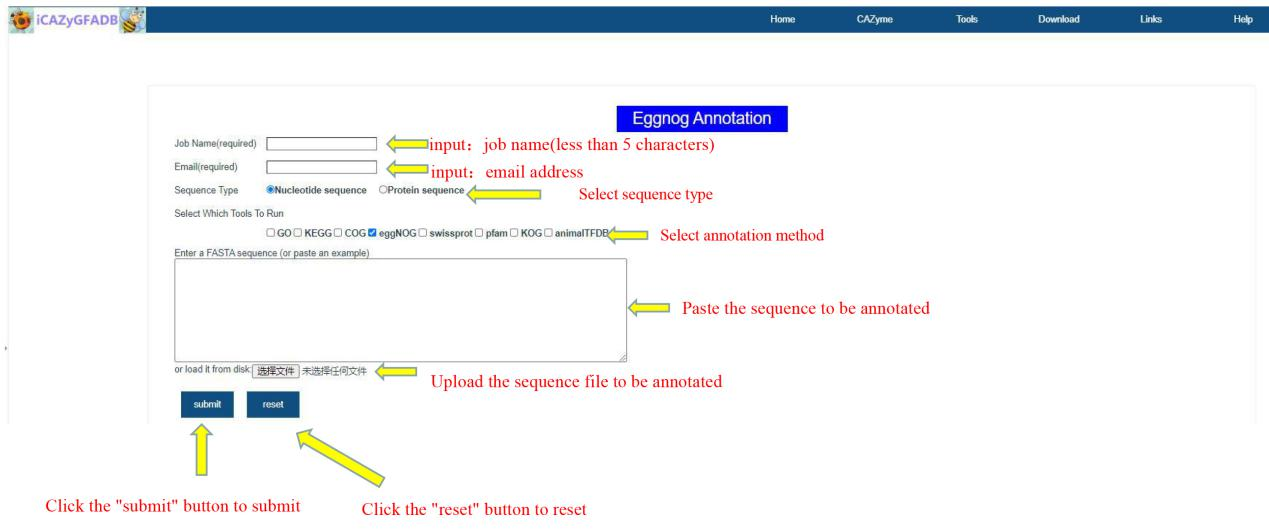
The first step is to input the job name (less than 5 characters) and email address. The second step is to select the sequence type(Nucleotide sequence OR Protein sequence) and annotation method. The third step is to upload the FASTA file of the annotation sequence. The fourth step is to submit.
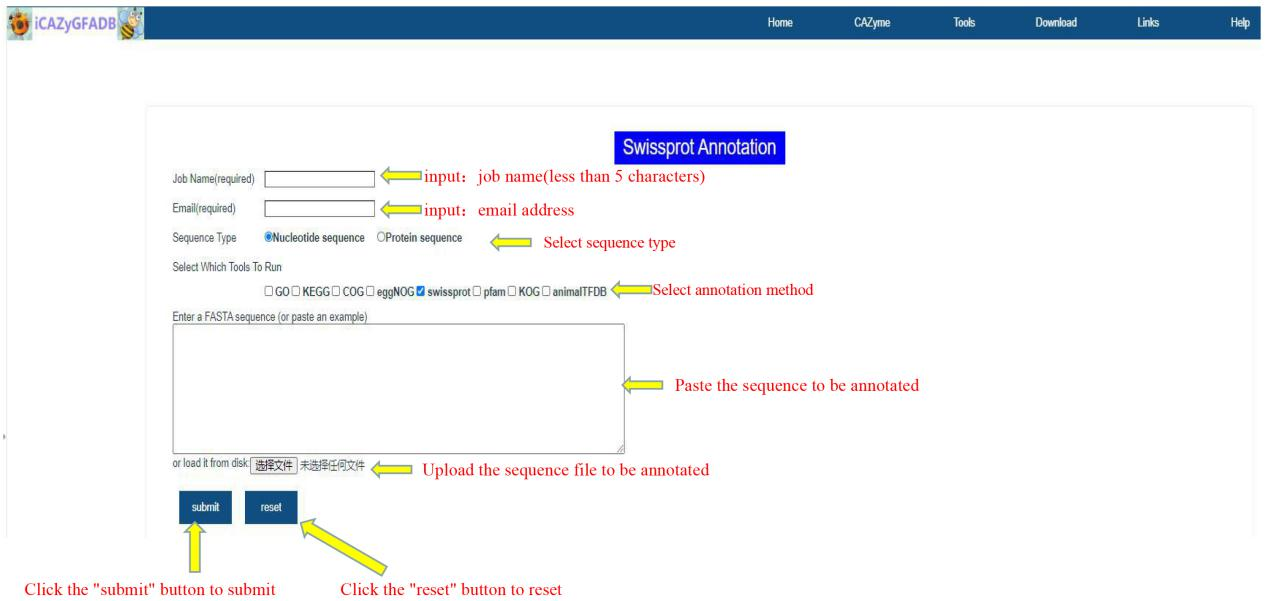
The first step is to input the job name (less than 5 characters) and email address. The second step is to select the sequence type(Nucleotide sequence OR Protein sequence) and annotation method. The third step is to upload the FASTA file of the annotation sequence. The fourth step is to submit.
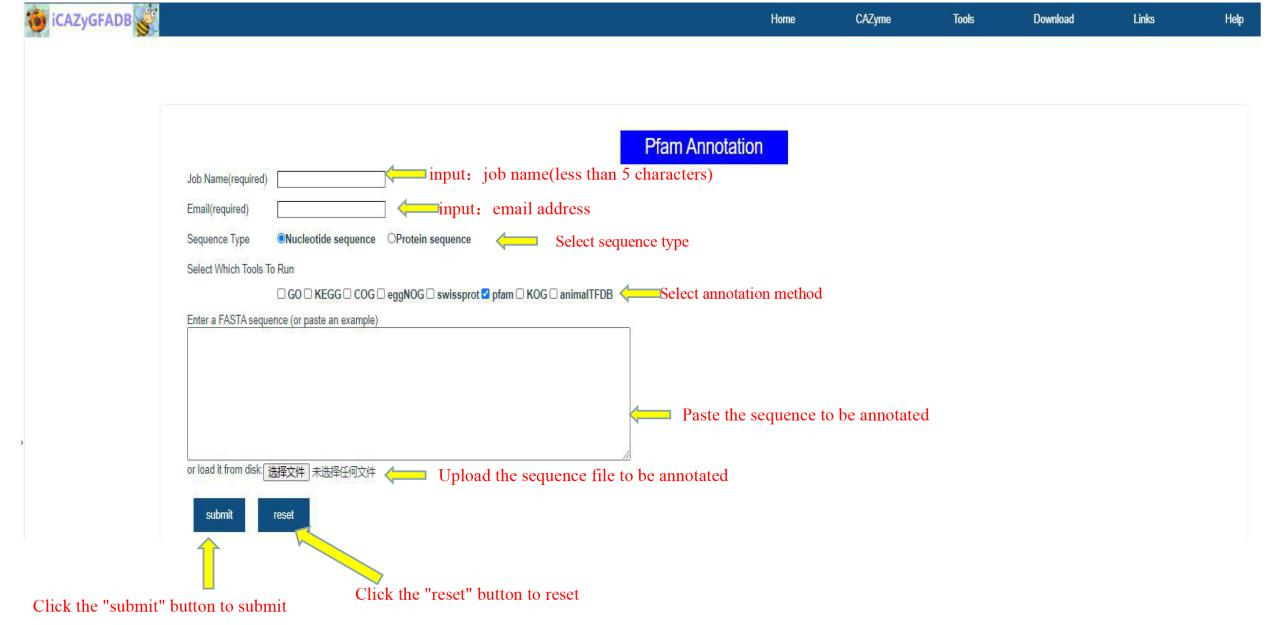
The first step is to input the job name (less than 5 characters) and email address. The second step is to select the sequence type(Nucleotide sequence OR Protein sequence) and annotation method. The third step is to upload the FASTA file of the annotation sequence. The fourth step is to submit.
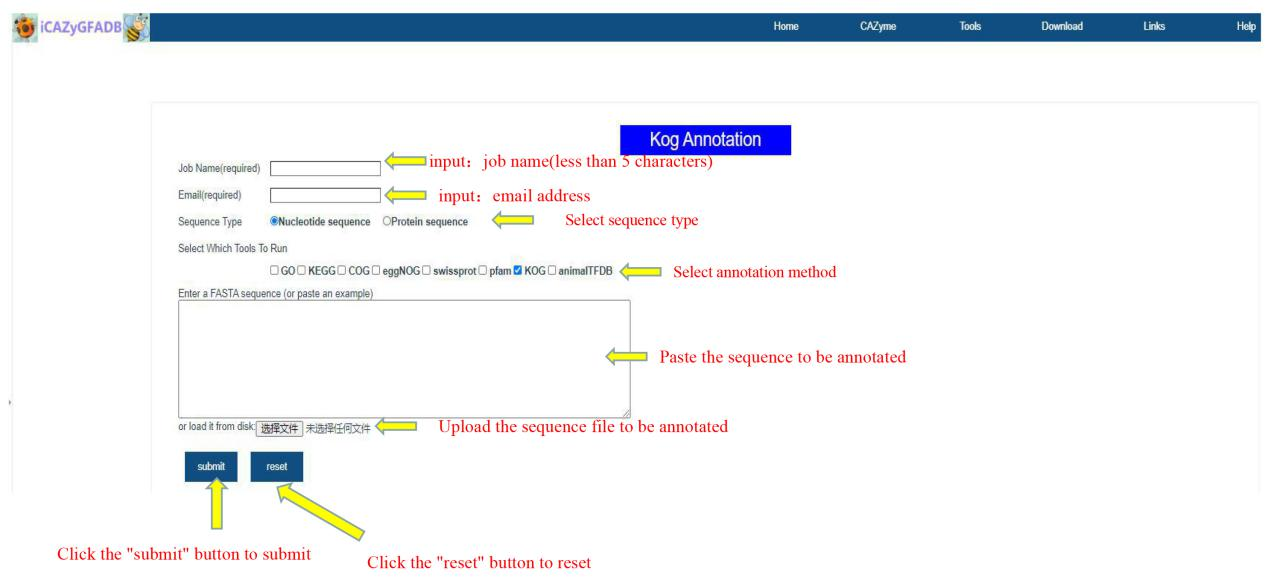
The first step is to input the job name (less than 5 characters) and email address. The second step is to select the sequence type(Nucleotide sequence OR Protein sequence) and annotation method. The third step is to upload the FASTA file of the annotation sequence. The fourth step is to submit.
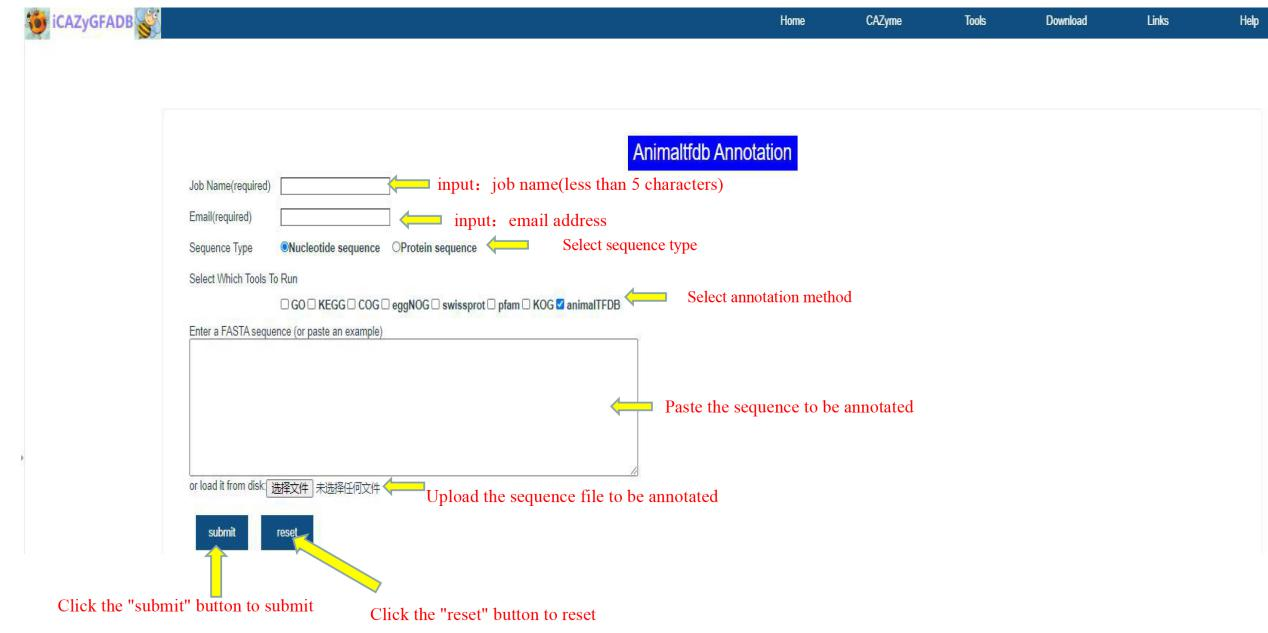
The first step is to input the job name (less than 5 characters) and email address. The second step is to select the sequence type(Nucleotide sequence OR Protein sequence), annotation method and reference species. The third step is to upload the FASTA file of the annotation sequence. The fourth step is to submit.

The download page provides CDS sequences and protein sequences of all members of CAZyme gene family in 151 insect genomes. The download page provides two download methods, namely, downloading from a single species and a single gene family. Just click download to download and save them to the local computer

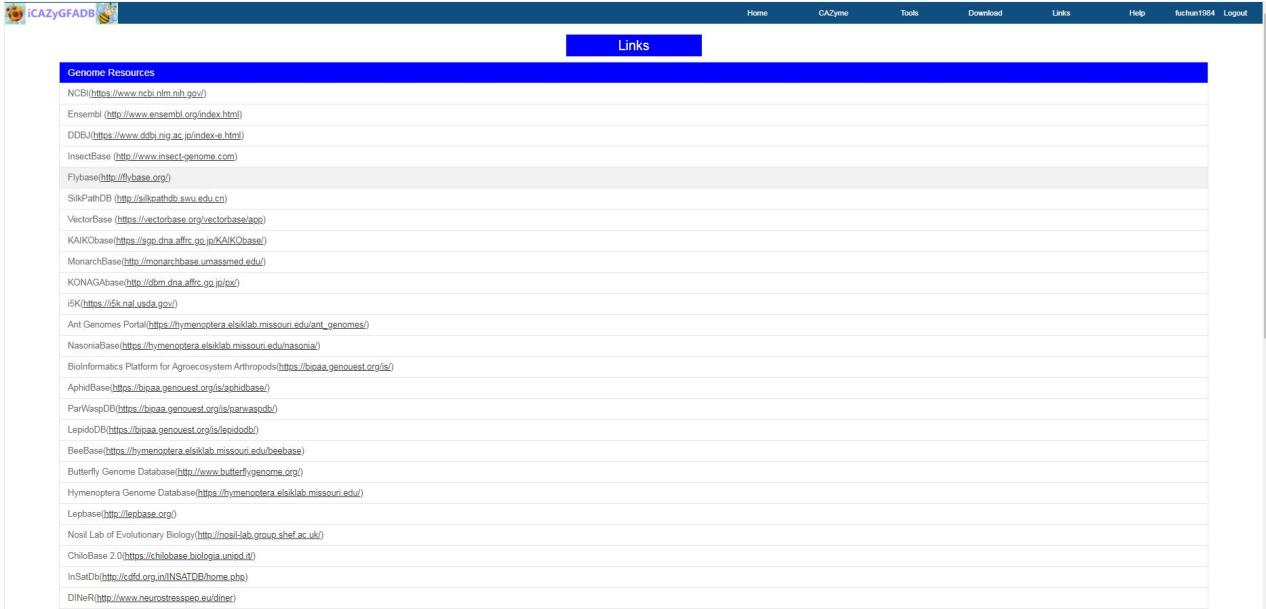
The links page contains two parts: Genome Resources and Genome annotation. The genomic data website links listed in Genome resources are the genomic data sources used in the construction of this database.The website links of gene function annotation tool listed in Genome annotation are the annotation tool sources used in gene function annotation tools of this database.

The help section introduces the operating instructions of this database website and provides a manual for users to use this database website.
This database website is completely open and free. All visiting scholars can register this database website. Users need to register an account before using the database website. When registering an account, they need to fill in the user name, password, institution, email and telephone.
After registering the account, the user can log in to the database website with the account name and password to use all annotation functions.